Introduction
In this post I will explore the concept of Cross Validation (CV) and its upgrade, the Nested Cross Validation, in a normal Machine Learning pipeline. It is often unclear when to use this fundamental technique and how to avoid information leakage.
This notebook was published on Kaggle and is part of the Song Popularity Competition in which I participated
Approach
When searching for parameters and estimating the error of a model, one could be tempted to use only one Cross Validation loop.
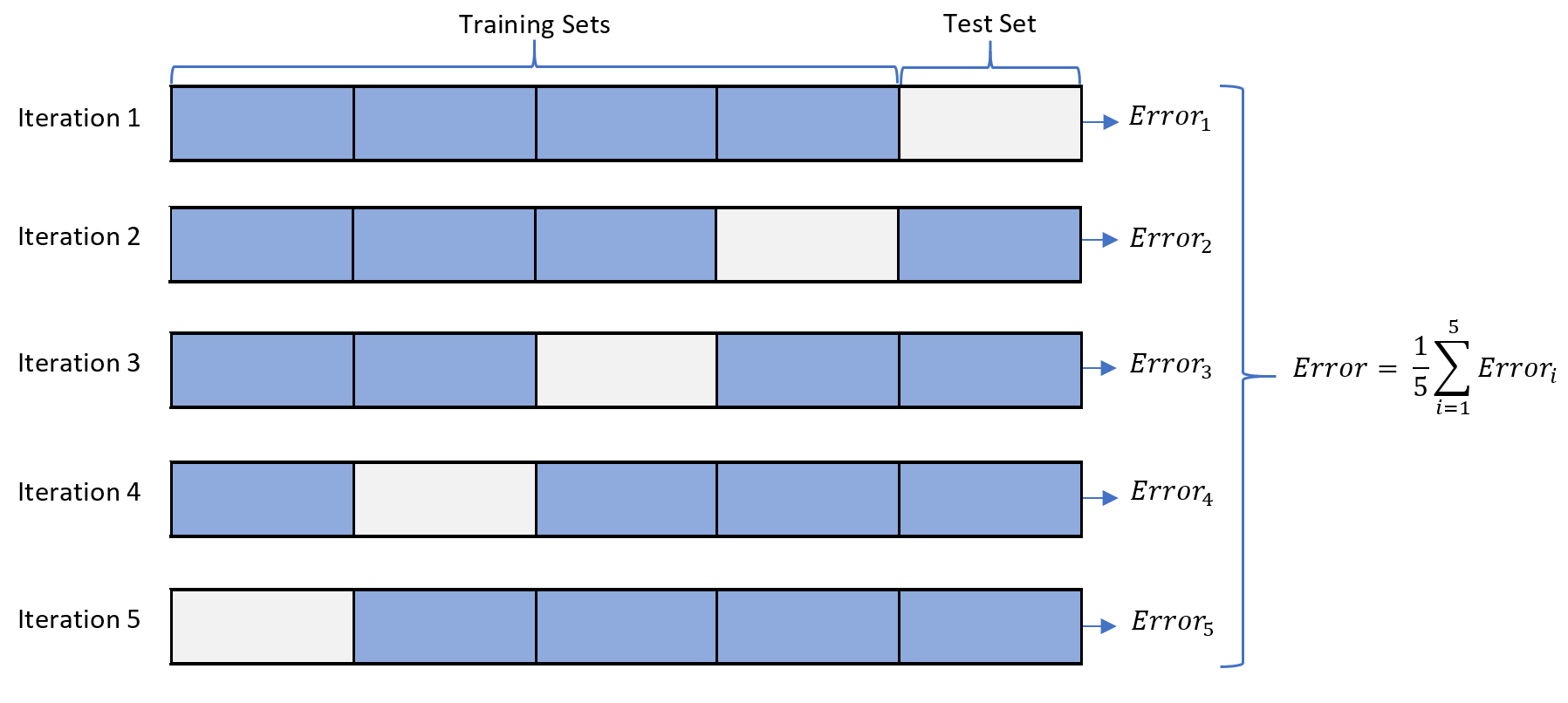
In the image above, we see an example of a generic K-fold cross validation. This procedure can be used both when optimizing the hyperparameters of a model and when comparing and selecting a model for the dataset. When the same cross-validation procedure and dataset are used to both tune and select a model, it is likely to lead to a biased evaluation of the model performance.
One way to avoid this is to set aside a piece of data, aka the test set.

In this approach:
- The dataset is divided in training, validation and test set
- The model is trained using the training fold
- Hyperparameters are tested on the validation set
- The final performance are evaluated on the test set.
What about Nested Cross Validation then?
To generalize better, we simply rotate the test set on the entire dataset. By doing so, we are sure to have a better overview of the performance of our model on the totality of the data.
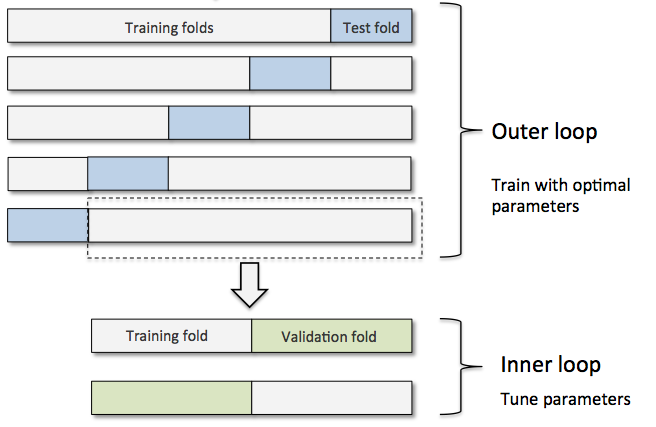
Let’s see it in action!
import numpy as np
import pandas as pd
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import StratifiedKFold, RandomizedSearchCV
from sklearn.metrics import auc, roc_auc_score
train = pd.read_csv('../input/song-popularity-prediction/train.csv').iloc[:, 1:]
test = pd.read_csv('../input/song-popularity-prediction/test.csv').iloc[:, 1:]
x = train.iloc[:, :-1]
y = train.song_popularity Framework
Let’s see first the code in action, then I’ll discuss it below
cv_inner = StratifiedKFold(n_splits=3, shuffle=True, random_state=42) #1
cv_outer = StratifiedKFold(n_splits=3, shuffle=True, random_state=42)
classifier = RandomForestClassifier(n_estimators=100, n_jobs=-1) #2
p_grid = {'min_samples_split': [60, 120, 180],
'max_depth': [5, 10, 15],
'max_features': ['sqrt', 'log2']}
history = []
pointer = 1
for train_index, test_index in cv_outer.split(x, y): #3
print('\nNestedCV: {} of outer fold {}'.format(pointer, cv_outer.get_n_splits()))
x_train, x_test = x.loc[train_index], x.loc[test_index]
y_train, y_test = y.loc[train_index], y.loc[test_index]
model = RandomizedSearchCV(classifier, param_distributions=p_grid,
scoring='roc_auc', cv=cv_inner, n_jobs=-1) #4
model.fit(x_train.fillna(-99), y_train)
pred_test = model.predict_proba(x_test.fillna(-99))
pred_training = model.predict_proba(x_train.fillna(-99))
auc_train = roc_auc_score(y_train, pred_training[:, 1]) #5
auc_test = roc_auc_score(y_test, pred_test[:, 1])
print("""
Best set of parameters: {}
Best AUC : {:.2f}
Training
AUC: {:.3f}
Test
AUC: {:.3f}
""".format(
model.best_params_,
model.best_score_,
auc_train,
auc_test,
)
)
history.append(auc_test)
pointer += 1
print('Overall test performance: {:.2f}'.format(np.mean(history)))NestedCV: 1 of outer fold 3
Best set of parameters: {'min_samples_split': 180, 'max_features': 'sqrt', 'max_depth': 10}
Best AUC : 0.57
Training
AUC: 0.718
Test
AUC: 0.567
NestedCV: 2 of outer fold 3
Best set of parameters: {'min_samples_split': 180, 'max_features': 'sqrt', 'max_depth': 5}
Best AUC : 0.56
Training
AUC: 0.612
Test
AUC: 0.566
NestedCV: 3 of outer fold 3
Best set of parameters: {'min_samples_split': 180, 'max_features': 'log2', 'max_depth': 10}
Best AUC : 0.57
Training
AUC: 0.721
Test
AUC: 0.565
Overall test performance: 0.57Code comment
#1.
cv_innerandcv_outerrepresent the external and internal CV splits of the NestedCV. Whilecv_outeris used to split the dataset in training/test,cv_innerwill be used by ourRandomizedSearchCVto find the best set of hyperparameters, splitting the training set in training/validation.#2. Here we init our base model and the grid parameters to search.
#3. Key moment, as we are generating the training/test folds
#4. As stated in #1,
cv_inneris being used byRandomizedSearchCVto find the best set of hyperparameters, splitting the training set in training/validation internally withcv=cv_inner#5. With the best set of hyperparameters, we then predict both on the training and test set to check the model performance
Results
As can be seen, the overall performance of the model is around 0.57, stable for all test splits generated.
The best parameters vary but overall we can say that we have a situation of model stability. For more reference about the concept of model stability, refer to this amazing answer on StackOverflow.